The run_command_file() plan – batch scans using a text file#
You can use a text file or an Excel spreadsheet as a multi-sample
batch scan tool using the run_command_file() plan.
This section is divided into these parts.
The Command File#
A command file can be written as either a plain text file or a spreadsheet (such as from Microsoft Excel or Libre Office).
Text Command File#
For example, given a text command file (named sample_example.txt)
with content as shown …
1# example text command file
2
3step_scan 5.07 8.3 0.0 "Water Blank, deionized"
4other_scan 5.07 8.3 0.0 "Water Blank, deionized"
5
6# all comment lines will be ignored
7
8this line will not be recognized by execute_command_list()
… can be summarized in a bluesky ipython session:
In [1]: import apstools.plans
In [2]: apstools.plans.summarize_command_file("sample_example.txt")
Command file: sample_example.xlsx
====== ========== ===========================================================
line # action parameters
====== ========== ===========================================================
3 step_scan 5.07, 8.3, 0.0, Water Blank, deionized
4 other_scan 5.07, 8.3, 0.0, Water Blank, deionized
8 this line, will, not, be, recognized, by, execute_command_list()
====== ========== ===========================================================
Observe which lines (3, 4, and 8) in the text file are part of
the summary. The other lines are either comments or blank.
The action on line 8 will be passed to execute_command_list() which will
then report the this action it is not handled.
(Looks like another type of a comment.)
See parse_text_command_file() for more details.
Spreadsheet Command File#
Follow the example spreadsheet (in the File Downloads for the Examples section)
and accompanying Jupyter notebook [1] to write your own Excel_plan().
For example, given a spreadsheet (named sample_example.xlsx)
with content as shown in the next figure:
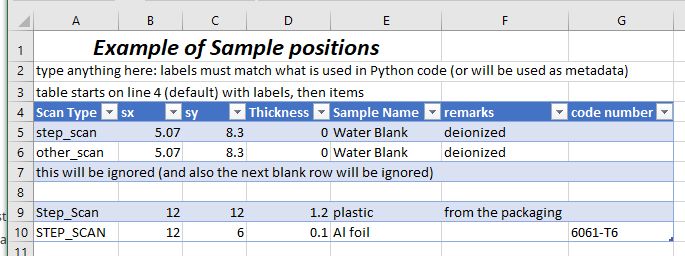
Image of sample_example.xlsx spreadsheet file.#
Tip
Place the column labels on the fourth row of the spreadsheet, starting in the first column. The actions start on the next row. The first blank row indicates the end of the command table within the spreadsheet. Use as many columns as you need, one column per argument.
This spreadsheet can be summarized in a bluesky ipython session:
In [1]: import apstools.plans
In [2]: apstools.plans.summarize_command_file("sample_example.xlsx")
Command file: sample_example.xlsx
====== ================================================================== ======================================
line # action parameters
====== ================================================================== ======================================
1 step_scan 5.07, 8.3, 0.0, Water Blank, deionized
2 other_scan 5.07, 8.3, 0.0, Water Blank, deionized
3 this will be ignored (and also the next blank row will be ignored)
====== ================================================================== ======================================
Note that lines 9 and 10 in the spreadsheet file are not part of
the summary. The spreadsheet handler will stop reading the list of actions
at the first blank line. The action described on line 3 will not be handled
(since we will not define an action named
this will be ignored (and also the next blank row will be ignored)).
See parse_Excel_command_file() for more details.
The Actions#
To use this example with the run_command_file() plan, it is
necessary to redefine the execute_command_list() plan,
we must write a plan to handle every different type of action described
in the spreadsheet. For sample_example.xlsx, we need to
handle the step_scan and other_scan actions.
Tip
Always write these actions as bluesky plans.
To test your actions, use either
bluesky.simulators.summarize_plan(step_scan())
or bluesky.simulators.summarize_plan(other_scan()).
Here are examples of those two actions (and a stub for an additional instrument procedure to make the example more realistic):
1def step_scan(sx, sy, thickness, title, md={}):
2 md["sample_title"] = title # log this metadata
3 md["sample_thickness"] = thickness # log this metadata
4 yield from bluesky.plan_stubs.mv(
5 sample_stage.x, sx,
6 sample_stage.y, sy,
7 )
8 yield from prepare_detector("scintillator")
9 yield from bluesky.plans.scan([scaler], motor, 0, 180, 360, md=md)
10
11def other_scan(sx, sy, thickness, title, md={}):
12 md["sample_title"] = title # log this metadata
13 md["sample_thickness"] = thickness # log this metadata
14 yield from bluesky.plan_stubs.mv(
15 sample_stage.x, sx,
16 sample_stage.y, sy,
17 )
18 yield from prepare_detector("areadetector")
19 yield from bluesky.plans.count([areadetector], md=md)
20
21def prepare_detector(detector_name):
22 # for this example, we do nothing
23 # we should move the proper detector into position here
24 yield from bluesky.plan_stubs.null()
Now, we replace execute_command_list()
with our own definition to include those two actions:
1def execute_command_list(filename, commands, md={}):
2 """our custom execute_command_list"""
3 full_filename = os.path.abspath(filename)
4
5 if len(commands) == 0:
6 yield from bps.null()
7 return
8
9 text = f"Command file: {filename}\n"
10 text += str(apstools.utils.command_list_as_table(commands))
11 print(text)
12
13 for command in commands:
14 action, args, i, raw_command = command
15 logger.info(f"file line {i}: {raw_command}")
16
17 _md = {}
18 _md["full_filename"] = full_filename
19 _md["filename"] = filename
20 _md["line_number"] = i
21 _md["action"] = action
22 _md["parameters"] = args # args is shorter than parameters, means the same thing here
23
24 _md.update(md or {}) # overlay with user-supplied metadata
25
26 action = action.lower()
27 if action == "step_scan":
28 yield from step_scan(*args, md=_md)
29 elif action == "other_scan":
30 yield from other_scan(*args, md=_md)
31
32 else:
33 logger.info(f"no handling for line {i}: {raw_command}")
Register our own execute_command_list#
Finally, we register our new version of execute_command_list
(which replaces the default execute_command_list()):
APS_plans.register_command_handler(execute_command_list)
If you wish to verify that your own code has been installed, use this command:
print(APS_plans._COMMAND_HANDLER_)
If its output contains apstools.plans.execute_command_list, you have not
registered your own function. However, if the output looks something such as
either of these:
<function __main__.execute_command_list(filename, commands, md={})>
# or
<function execute_command_list at 0x7f4cf0d616a8>
then you have installed your own code.
Testing the command file#
As you were developing plans for each of your actions, we showed you
how to test that each plan was free of basic syntax and bluesky procedural
errors. The bluesky.simulators.summarize_plan() function
will run through your plan and show you the basic data acquisition steps
that will be executed during your plan. Becuase you did not write
any blocking code, no hardware should ever be changed by running
this plan summary.
To test our command file, run it through the
bluesky.simulators.summarize_plan() function:
bluesky.simulators.summarize_plan(run_command_file("sample_example.txt"))
The output will be rather lengthy, if there are no errors. Here are the first few lines:
Command file: sample_example.txt
====== ========== ===========================================================
line # action parameters
====== ========== ===========================================================
3 step_scan 5.07, 8.3, 0.0, Water Blank, deionized
4 other_scan 5.07, 8.3, 0.0, Water Blank, deionized
8 this line, will, not, be, recognized, by, execute_command_list()
====== ========== ===========================================================
file line 3: step_scan 5.07 8.3 0.0 "Water Blank, deionized"
sample_stage_x -> 5.07
sample_stage_y -> 8.3
=================================== Open Run ===================================
m3 -> 0.0
Read ['scaler', 'm3']
m3 -> 0.5013927576601671
Read ['scaler', 'm3']
m3 -> 1.0027855153203342
Read ['scaler', 'm3']
m3 -> 1.5041782729805013
and the last few lines:
m3 -> 178.99721448467966
Read ['scaler', 'm3']
m3 -> 179.49860724233983
Read ['scaler', 'm3']
m3 -> 180.0
Read ['scaler', 'm3']
================================== Close Run ===================================
file line 4: other_scan 5.07 8.3 0.0 "Water Blank, deionized"
sample_stage_x -> 5.07
sample_stage_y -> 8.3
=================================== Open Run ===================================
Read ['areadetector']
================================== Close Run ===================================
file line 8: this line will not be recognized by execute_command_list()
no handling for line 8: this line will not be recognized by execute_command_list()
Running the command file#
Prepare the RE#
These steps were used to prepare our bluesky ipython session to run the plan:
1from bluesky import RunEngine
2from bluesky.utils import get_history
3RE = RunEngine(get_history())
4
5# Import matplotlib and put it in interactive mode.
6import matplotlib.pyplot as plt
7plt.ion()
8
9# load config from ~/.config/databroker/mongodb_config.yml
10from databroker import Broker
11db = Broker.named("mongodb_config")
12
13# Subscribe metadatastore to documents.
14# If this is removed, data is not saved to metadatastore.
15RE.subscribe(db.insert)
16
17# Set up SupplementalData.
18from bluesky import SupplementalData
19sd = SupplementalData()
20RE.preprocessors.append(sd)
21
22# Add a progress bar.
23from bluesky.utils import ProgressBarManager
24pbar_manager = ProgressBarManager()
25RE.waiting_hook = pbar_manager
26
27# Register bluesky IPython magics.
28from bluesky.magics import BlueskyMagics
29get_ipython().register_magics(BlueskyMagics)
30
31# Set up the BestEffortCallback.
32from bluesky.callbacks.best_effort import BestEffortCallback
33bec = BestEffortCallback()
34RE.subscribe(bec)
Also, since we are using an EPICS area detector (ADSimDetector) and have just
started its IOC, we must process at least one image from the CAM to each of the
file writing plugins we’ll use (just the HDF1 for us). A procedure has been added
to the ophyd.areadetector code for this. Here is the command we used
for this procedure:
areadetector.hdf1.warmup()
Run the command file#
To run the command file, you need to pass this to an instance of the
bluesky.RunEngine, defined as RE above:
RE(apstools.plans.run_command_file("sample_example.txt"))
The output will be rather lengthy. Here are the first few lines of the output on my system (your hardware may be different so the exact data columns and values will vary):
In [11]: RE(APS_plans.run_command_file("sample_example.txt"))
Command file: sample_example.txt
====== ========== ===========================================================
line # action parameters
====== ========== ===========================================================
3 step_scan 5.07, 8.3, 0.0, Water Blank, deionized
4 other_scan 5.07, 8.3, 0.0, Water Blank, deionized
8 this line, will, not, be, recognized, by, execute_command_list()
====== ========== ===========================================================
file line 3: step_scan 5.07 8.3 0.0 "Water Blank, deionized"
Transient Scan ID: 138 Time: 2019-07-04 16:52:15
Persistent Unique Scan ID: 'f40d1c3c-8361-4efc-83f1-072fcd44c1b2'
New stream: 'primary'
+-----------+------------+------------+------------+------------+------------+
| seq_num | time | m3 | clock | I0 | scint |
+-----------+------------+------------+------------+------------+------------+
| 1 | 16:52:34.5 | 0.00000 | 4000000 | 2 | 1 |
| 2 | 16:52:35.2 | 0.50000 | 4000000 | 1 | 2 |
| 3 | 16:52:36.0 | 1.00000 | 4000000 | 1 | 1 |
| 4 | 16:52:36.6 | 1.50000 | 4000000 | 2 | 1 |
| 5 | 16:52:37.3 | 2.01000 | 4000000 | 1 | 2 |
| 6 | 16:52:38.0 | 2.51000 | 4000000 | 2 | 2 |
| 7 | 16:52:38.7 | 3.01000 | 4000000 | 1 | 1 |
| 8 | 16:52:39.3 | 3.51000 | 4000000 | 1 | 2 |
and the last few lines:
| 352 | 16:56:53.4 | 175.99000 | 4000000 | 3 | 0 |
| 353 | 16:56:54.1 | 176.49000 | 4000000 | 3 | 1 |
| 354 | 16:56:55.0 | 176.99000 | 4000000 | 1 | 3 |
| 355 | 16:56:55.7 | 177.49000 | 4000000 | 2 | 1 |
| 356 | 16:56:56.4 | 177.99000 | 4000000 | 1 | 1 |
| 357 | 16:56:57.1 | 178.50000 | 4000000 | 2 | 1 |
| 358 | 16:56:57.8 | 179.00000 | 4000000 | 2 | 2 |
| 359 | 16:56:58.5 | 179.50000 | 4000000 | 1 | 2 |
| 360 | 16:56:59.2 | 180.00000 | 4000000 | 2 | 1 |
+-----------+------------+------------+------------+------------+------------+
generator scan ['f40d1c3c'] (scan num: 138)
file line 4: other_scan 5.07 8.3 0.0 "Water Blank, deionized"
Transient Scan ID: 139 Time: 2019-07-04 16:56:59
Persistent Unique Scan ID: '1f093f56-3413-4e67-8a1b-27e71881b855'
New stream: 'primary'
+-----------+------------+
| seq_num | time |
+-----------+------------+
| 1 | 16:56:59.7 |
+-----------+------------+
generator count ['1f093f56'] (scan num: 139)
file line 8: this line will not be recognized by execute_command_list()
no handling for line 8: this line will not be recognized by execute_command_list()
Out[11]:
('f40d1c3c-8361-4efc-83f1-072fcd44c1b2',
'1f093f56-3413-4e67-8a1b-27e71881b855')
In [12]:
Appendix: Other spreadsheet examples#
You can use an Excel spreadsheet as a multi-sample batch scan tool.
SIMPLE: Your Excel spreadsheet could be rather simple…
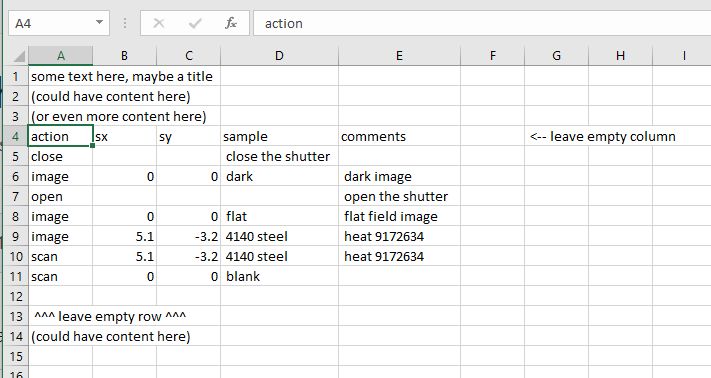
Unformatted Excel spreadsheet for batch scans.#
See ExcelDatabaseFileGeneric for an example bluesky plan
that reads from this spreadsheet.
FANCY: … or contain much more information, including formatting.
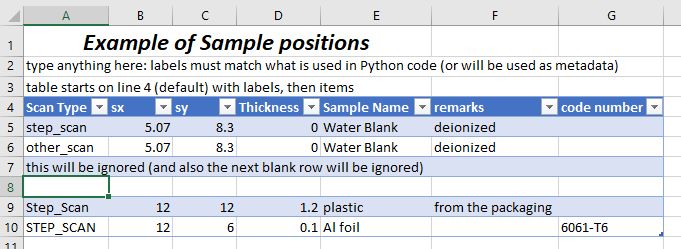
Example Excel spreadsheet for multi-sample batch scans.#
The idea is that your table will start with column labels
in row 4 of the Excel spreadsheet. One of the columns will be the name
of the action (in the example, it is Scan Type). The other columns will
be parameters or other information. Each of the rows under the labels will
describe one type of action such as a scan. Basically, whatever you
handle in your Excel_plan().
Any rows that you do not handle will be reported to the console during execution
but will not result in any action.
Grow (or shrink) the table as needed.
Note
For now, make sure there is no content in any of the spreadsheet cells outside (either below or to the right) of your table. Such content will trigger a cryptic error about a numpy float that cannot be converted. Instead, put that content in a second spreadsheet page.
See also
You’ll need to have an action plan for every different action your spreadsheet
will specify. Call these plans from your Excel_plan() within an elif block,
as shown in this example. The example Excel_plan() converts the Scan Type
into lower case for simpler comparisons. Your plan can be different if you choose.
if scan_command == "step_scan":
yield from step_scan(...)
elif scan_command == "energy_scan":
yield from scan_energy(...)
elif scan_command == "radiograph":
yield from AcquireImage(...)
else:
print(f"no handling for table row {i+1}: {row}")
The example plan saves all row parameters as metadata to the row’s action. This may be useful for diagnostic purposes.