The lineup() plan - align an axis with a signal#
In this example, we demonstrate the apstools.plans.lineup() plan, which aligns
an axis using some statistical measure (cen: centroid, com: center of mass,
max: position of peak value, or even min: negative trending peaks) of a
signal. If an alignment is possible, an optional rescan will fine-tune the
alignment within the width and center of the first scan. Use rescan=False
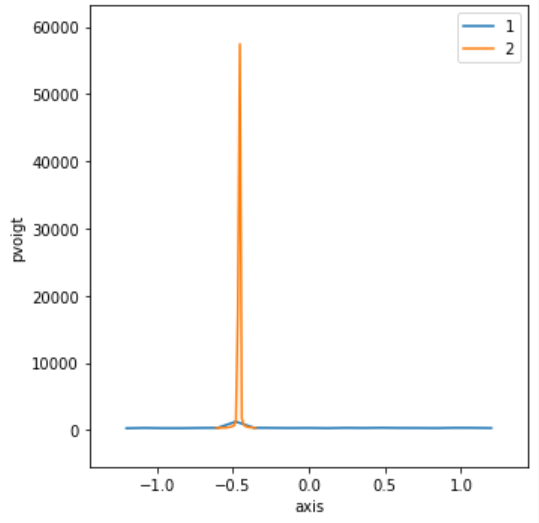
keyword to disable. Here’s an example chart showing the first (roughly, locate
at least one point within the peak) and second (fine-tune the position)
scans.
We’ll use a floating-point scalar value (not connected to hardware) as a positioner. Then, we prepare a simulated detector signal that is a computation based on the value of our positioner. The computed signal is a model of a realistic diffraction peak (pseudo-Voigt, a mixture of a Gaussian and a Lorentzian) one might encounter in a powder diffraction scan. The model peak is a pseudo-voigt function to which some noise has been added. Random numbers are used to modify the ideal pseudo-voigt function so as to simulate a realistic signal.
For this demo, we’ll use a temporary databroker catalog (deleted when the
notebook is closed) since we do not plan to review any of this data after
collection. We’ll display the data during the scan(in both a table and a chart)
using a BestEffortCallback() subscription to the bluesky.RunEngine().
from apstools.devices import SynPseudoVoigt
from apstools.plans import lineup
from bluesky import RunEngine
from bluesky.callbacks import best_effort
import bluesky.plan_stubs as bps
import databroker
import numpy
import ophyd
bec = best_effort.BestEffortCallback()
cat = databroker.temp()
RE = RunEngine({})
RE.subscribe(cat.v1.insert)
RE.subscribe(bec)
1
Setup#
Set the IOC prefix and connect with our EPICS PVs.
IOC = "gp:"
axis = ophyd.EpicsSignal(f"{IOC}gp:float1", name="axis")
axis.wait_for_connection()
Once connected, create the detector signal (the computed pseudo-Voigt) with default peak parameters.
# Need to know that axis is connected before using here.
pvoigt = SynPseudoVoigt(name="pvoigt", motor=axis, motor_field=axis.name)
The pvoigt signal must have kind="hinted" for it to appear in tables and plots.
pvoigt.kind = "hinted"
Move axis to a starting position. Pick zero.
RE(bps.mv(axis, 0))
()
Scan#
To make things interesting, first randomize the peak parameters. (Peak is placed randomly between -1..+1 on axis scale, with random width, scale, pseudo-Voigt mixing parameter, noise, …)
pvoigt.randomize_parameters(scale=100_000)
Run the lineup() plan through the range where the peak is expected. Don’t need many points to catch some value that is acceptable (max is more than 4*min) the background.
RE(lineup(pvoigt, axis, -1.2, 1.2, 13, feature="cen", rescan=True))
print(f"{pvoigt.read()=}, {axis.get()=}")
Transient Scan ID: 1 Time: 2022-09-28 15:35:18
Persistent Unique Scan ID: '22182d47-72c1-450b-bf62-035a2c608d11'
New stream: 'primary'
+-----------+------------+------------+------------+
| seq_num | time | axis | pvoigt |
+-----------+------------+------------+------------+
| 1 | 15:35:18.6 | -1.2000 | 1413 |
| 2 | 15:35:18.6 | -1.0000 | 5259 |
| 3 | 15:35:18.7 | -0.8000 | 55856 |
| 4 | 15:35:18.7 | -0.6000 | 2113 |
| 5 | 15:35:18.7 | -0.4000 | 1070 |
| 6 | 15:35:18.8 | -0.2000 | 796 |
| 7 | 15:35:18.8 | 0.0000 | 689 |
| 8 | 15:35:18.8 | 0.2000 | 675 |
| 9 | 15:35:18.8 | 0.4000 | 600 |
| 10 | 15:35:18.8 | 0.6000 | 649 |
| 11 | 15:35:18.9 | 0.8000 | 567 |
| 12 | 15:35:18.9 | 1.0000 | 547 |
| 13 | 15:35:18.9 | 1.2000 | 552 |
+-----------+------------+------------+------------+
generator rel_scan ['22182d47'] (scan num: 1)
Transient Scan ID: 2 Time: 2022-09-28 15:35:19
Persistent Unique Scan ID: '9928a319-fa14-4b5e-837f-1f4058f7029c'
New stream: 'primary'
+-----------+------------+------------+------------+
| seq_num | time | axis | pvoigt |
+-----------+------------+------------+------------+
| 1 | 15:35:19.1 | -1.0154 | 4403 |
| 2 | 15:35:19.1 | -0.9801 | 7746 |
| 3 | 15:35:19.1 | -0.9447 | 18831 |
| 4 | 15:35:19.2 | -0.9093 | 46123 |
| 5 | 15:35:19.2 | -0.8739 | 88448 |
| 6 | 15:35:19.2 | -0.8386 | 99186 |
| 7 | 15:35:19.2 | -0.8032 | 59675 |
| 8 | 15:35:19.2 | -0.7678 | 25649 |
| 9 | 15:35:19.3 | -0.7325 | 10329 |
| 10 | 15:35:19.3 | -0.6971 | 5200 |
| 11 | 15:35:19.3 | -0.6617 | 3386 |
| 12 | 15:35:19.3 | -0.6263 | 2602 |
| 13 | 15:35:19.3 | -0.5910 | 2046 |
+-----------+------------+------------+------------+
generator rel_scan ['9928a319'] (scan num: 2)
pvoigt.read()={'pvoigt': {'value': 2046, 'timestamp': 1664397319.3793561}}, axis.get()=-0.8496701712914874
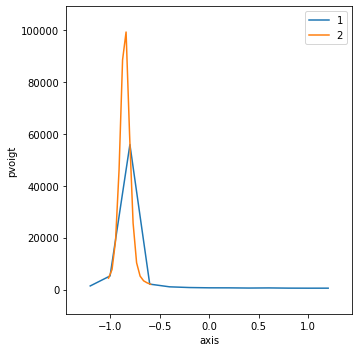
Validate#
Show the position after the lineup() completes. Test (Python assert) that it is within the expected range.
center = pvoigt.center
sigma = 2.355 * pvoigt.sigma
print(f"{center=}\n{sigma=}\n{bec.peaks=}")
assert center-sigma <= axis.get() <= center+sigma
center=-0.8503901139652472
sigma=0.11456520969899238
bec.peaks={
'com':
{'pvoigt': -0.8461974129441658}
,
'cen':
{'pvoigt': -0.8496701712914874}
,
'max':
{'pvoigt': (-0.8385705807871623,
99186)}
,
'min':
{'pvoigt': (-0.5909727956676526,
2046)}
,
'fwhm':
{'pvoigt': 0.11177564414922392}
,
}